Biomedical Research Assistant
Biomedical Research Assistant built using RAG, LangChain, and LLaMA3
Read on MediumI am an AI/ML and Bioinformatics professional with 3 years of experience, specialized in building scalable and robust AI/ML models, bioinformatics pipelines, and data analysis tools, focusing on advancing drug discovery, precision medicine, target identification, clinical development and therapeutic research through cross-functional collaboration and data-driven solutions. Passionate about solving complex biomedical problems at the intersection of AI, omics, and healthcare. Gmail | LinkedIN | GitHub
Multi disciplinary capabilities across research, therapeutics, and technology development
Discovering and validating therapeutic targets
Designing potential therapeutic candidates
Evaluating safety and efficacy
Extracting biological insights from complex data
Understanding disease mechanisms
Bridging research to patient care
Ingesting and standardizing biological data
Building robust analysis workflows
Deriving actionable insights
Structuring biomedical data for AI
Building domain specific AI solutions
Implementing solutions in practice
Developed an AI driven pipeline for antibody sequence generation, leveraging deep learning and generative models trained on large scale OAS & SAbDab antibody datasets. Integrated with sequence optimization and structural validation tools to design high affinity, developable antibodies with therapeutic potential.
AntiFold | AbLang | diffAb | BioPhi
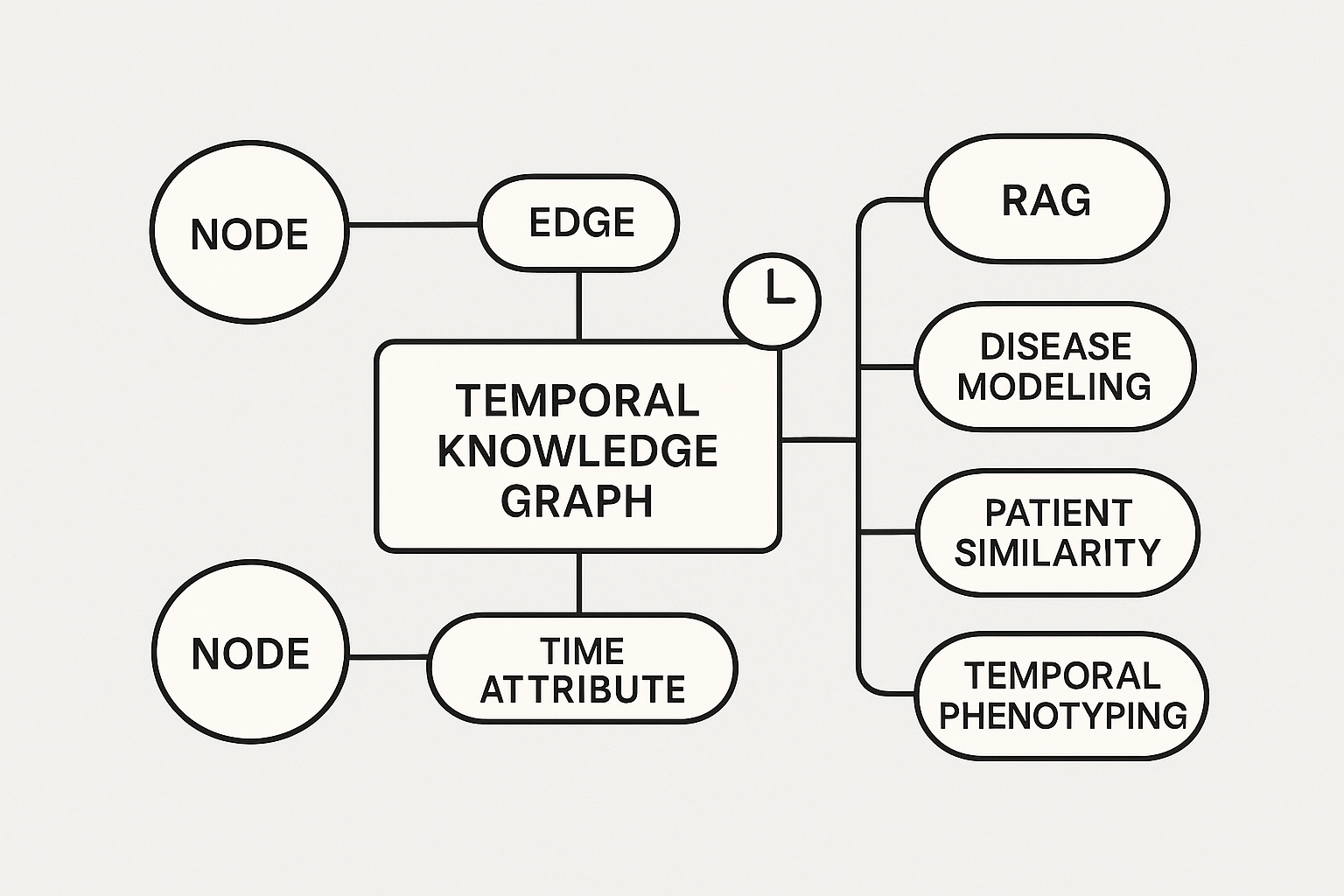
Built a KG with open source biological/Real World/Clinical data, harmonized with controlled vocabularies for each entity. Application included drug repurposing, target identification, safety assessment for toxicity and organ wise stratification, reducing months of work to weeks
Neo4j | Link prediction | Node classification | Community Detection
Developed an automated ML pipeline to build Quantitative Structure-Property Relationship (QSPR) model for drug property prediction, that helped reducing dependency on data scientist for model building and increased capabilities across departments.
RDKit | Morgen fingerprint | Optuna - Bayesian Optimization
Developed a computational workflow for biomarker identification, by training ensemble ML models using omics data (RNA seq, scRNA seq), to identify diagnostic and prognostic signatures, enabling patient stratification for precision medicine applications.
XGBoost | Feature importance | ROC | Pathway enrichment
1. GenomeIndia cohort: Quality control and variant association (SNPs) studies.
2. AllOfUs cohort: Developed a workflow for loss of function association studies to identify biomarkers with the help of recorded EHR data.
Association statistics | Dosage sensitivity | Burden test
Developed a high throughput Boolean model simulation pipeline for in silico gene knockout/perturbation experiments, using RNA seq data to initialize the system states supporting data-driven therapeutics which enhances precision in target prioritization.
Attractors | Trap spaces | Logical gates | KEGG
Developed a structure based druggability prediction pipeline leveraging parallel processing to accelerate searches across a database of known binding pockets, enabling rapid identification of similar sites to assess target protein druggability.
Graph Analytics | Structural bioinformatics | Multiprocessing
Integrative data analysis (IDA) of Breast cancer (BRCA) dataset for predictive model.
Predictive models for drug toxicity using data from tox24 challange
MedMNIST datasets was used for building classification models(CNN) for different modalities
Coming soon !

Biomedical Research Assistant built using RAG, LangChain, and LLaMA3
Read on Medium